Constructs
Nup42, Nup50, Nup54, Nup62, Nup88, Nup133 and Nup214 in the pDONR221 gateway master vector were purchased from DNASU and cloned into the cell=free expression vector pCellFree_03 (ref. 55), using the Gateway method, to produce fusion proteins with an N-terminal GFP tag (see Supplementary Table 2 for details). Nup35, Nup58, Nup98, Nup358 and Pom121 were cloned from complementary DNA (cDNA) prepared from HEK293T cells. Briefly, RNA was extracted using an Isolate II RNA mini kit (Bioline), and cDNA was synthesized using oligo(dT) primers and SuperScript IV (Thermo Fisher Scientific). Target genes were then cloned into pCellFree_03 using the Gibson assembly protocol (New England Biolabs). Nup98 and Pom121 fragments were purchased as gBlocks from IDT and cloned into pCellFree_03. pET28a vectors containing mCherry2-Nup98 fragments, as well as pGEX-6P-3 vectors containing importin-α, TNPO1 and TNPO3 were purchased from GenScript. Cell lines (HEK239T) were only used for cDNA preparation in this study, with gene identity subsequently confirmed by Sanger sequencing.
Protein expression and purification
HIV-1 CA proteins
HIV-1 CA proteins (K158C, A204C, R18G/A204C, N57D/A204C, N74D/A204C, N77V/A204C) were expressed in Escherichia coli C41 or Rosetta2 cells and purified as previously described56. HIV-1 CA K158C was labelled with Alexa Fluor 568-C5-maleimide (AF568, Thermo Fisher Scientific, A20341) and mixed in an approximately 1:200 ratio with unlabelled HIV-1 CA A204C before assembly. CA lattice assembly was carried out as described by Lau et al. 17 with the modification of a 15 min incubation at 37 °C and no overnight incubation. CAhexamer, CAhexamer-mCherry and CA-mCherry were purified based on previously described protocols27. Briefly, CAhexamer, CAhexamer-mCherry (pOPT) and CA-mCherry (pET21a) were expressed in E. coli C41 cells. Cells were grown at 37 °C, protein expression was induced with 0.5 mM isopropyl β-d-1-thiogalactopyranoside (IPTG) at an optical density of 0.6, and cell growth was continued overnight at 18 °C. Cells were collected (4,000g, 10 min, 4 °C), and the cell pellets were resuspended in lysis buffer (purification buffer 50 mM Tris pH 8, 50 mM NaCl, 2 mM dithiothreitol (DTT); +cOmplete EDTA-free Protease Inhibitor (Roche)) and lysed by sonication. The lysate was cleared by centrifugation (16,000g, 60 min, 4 °C), and an ammonium sulfate (20% w/v) precipitation was carried out on the soluble fraction. After stirring for 30 min at 4 °C, the precipitated material was pelleted (16,000g, 20 min, 4 °C). CAhexamer was further purified by resuspending the pellet in 100 mM citric acid pH 4.5, 2 mM DTT and dialysed against the same buffer three times. The precipitated protein was pelleted (16,000g, 20 min, 4 °C), and the soluble fraction was collected for further purification. CAhexamer-mCherry and CA-mCherry could not be purified with citric acid precipitation owing to a possible loss of mCherry fluorescence. The ammonium-sulfate-precipitated pellets were resuspended in 8 ml purification buffer per litre culture, run over a 20 ml Hi-TRAP Q column in purification buffer and eluted in a 1–100% gradient with buffer B (50 mM Tris pH 8, 500 mM NaCl). Protein-containing fractions were pooled.
CA-mCherry was further purified by gel filtration (Superdex 200 in purification buffer), and fractions containing clean protein were pooled and snap frozen to be stored at −80 °C.
To assemble the final cross-linked hexameric CAhexamer-mCherry, CAhexamer and CAhexamer-mCherry were mixed in a 5:1 ratio and assembled by means of the following dialysis steps: twice against 50 mM Tris pH 8, 1,000 mM NaCl, 2 mM DTT; twice against 50 mM Tris pH 8, 1,000 mM NaCl; and twice against 50 mM Tris pH 8, 40 mM NaCl. The assembled CAhexamer-mCherry was finally purified with gel filtration (S200), and fractions containing hexameric CAhexamer-mCherry carrying one mCherry were pooled and snap frozen to be stored at −80 °C.
Cell-free expression
Cell-free expression of GFP fusion proteins was performed as described by Lau et al.17. Briefly Leishmania tarentolae extract was supplemented with RnaseOUT (1:1,000, Invitrogen) on ice. pCellFree_03-Nup plasmids (150 ng μl−1) were then added to the expression mix, followed by incubation at 27 °C for 3 h. Following incubation, the undiluted extract was mixed 1:1 with NuPAGE LDS loading buffer (Thermo Fisher Scientific), 10 mM DTT, and the purity and degradation of the expressed Nup were assessed by sodium dodecyl sulfate polyacrylamide gel electrophoresis in-gel fluorescence.
Importin-β, IBB-mCherry, Nup98
The mCherry protein was kindly provided by J. Goyette. Importin-β and IBB-mCherry in pET11a (GenScript) were expressed as His-fusion proteins in E. coli Rosetta2 cells. Cells were grown at 37 °C, protein expression was induced with 0.5 mM IPTG at an optical density of 0.6, and cell growth was continued overnight at 18 °C. Cells were collected (4,000g, 10 min, 4 °C), and the cell pellets were resuspended in lysis buffer (purification buffer: 20 mM Tris pH 7.5, 150 mM NaCl, 2 mM DTT; +cOmplete EDTA-free Protease Inhibitor (Roche)) and lysed by sonication. The lysate was cleared by centrifugation (16,000g, 45 min, 4 °C), and the supernatant was bound to Ni-beads (Ni Sepharose High Performance, Cytiva) preincubated in purification buffer; then, imidazole was added to the protein–Ni slurry to a final concentration of 10 mM, and the mixture was incubated for 2 h at 4 °C. The beads were washed with 20 column volumes of wash buffer (purification buffer + 20 mM imidazole) and eluted in 2 ml fractions with elution buffer (purification buffer + 500 mM imidazole). Protein-containing fractions were pooled, and cleavage of the His-tag was achieved with TEV (IBB-mCherry) or SUMO (importin-β) in overnight dialysis against TEV-cleavage buffer (50 mM Tris pH 7.5, 1 mM DTT) or SUMO-cleavage buffer (50 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT). Cleaved proteins were added to Ni-beads, and the flow-through was collected. Proteins were finally purified by size-exclusion chromatography (SEC; Hi Load16/60 Superdex 200) in purification buffer. Clean protein fractions were pooled and snap frozen to be stored at −80 °C. Nup98 in pET28a was purified as described above with the following changes: the buffers used were purification buffer (6 M GuHCl, 50 mM TrisHCl pH 7.5, 2 mM DTT), wash buffer (6 M GuHCl, 20 mM imidazole pH 8) and elution buffer (500 mM GuHCl, 500 mM imidazole pH 8). After induction with IPTG, cells were grown further for 3 h at 30 °C before being collected. Sufficient purity of the protein was achieved with affinity chromatography; consequently, no SEC was performed. The His-tag was not removed. The protein–Ni slurry was stored at 4 °C for phase-separation assays.
Importin-α, TNPO1, TNPO3
Importin-α, TNPO1 and TNPO3 were expressed as GST-fusion proteins from E. coli Rosetta2 cells, as described above for importin-β and IBB-mCherry. Cell pellets were then resuspended in 25 mM Tris pH 7.5, 500 mM NaCl, 1 mM DTT, 10% glycerol and 1× cOmplete EDTA-free Protease Inhibitor (Roche) and lysed by sonication. Cell lysates were subjected to centrifugation (18,000g, 60 min, 4 °C). Clarified lysates were bound to GSH-resin and washed with 25 mM Tris pH 7.5, 150 mM NaCl, 1 mM DTT, 10% glycerol. On-column cleavage of the GST-tag was achieved with PreScission Protease. Cleaved proteins were subjected to SEC over a Superdex 200 26/600 (Cytiva) and equilibrated in 50 mM Tris pH 7.5, 200 mM NaCl, 1 mM DTT, 10% glycerol. Eluted proteins were concentrated, snap frozen and stored at – 80 °C.
Fluorescence fluctuation spectroscopy
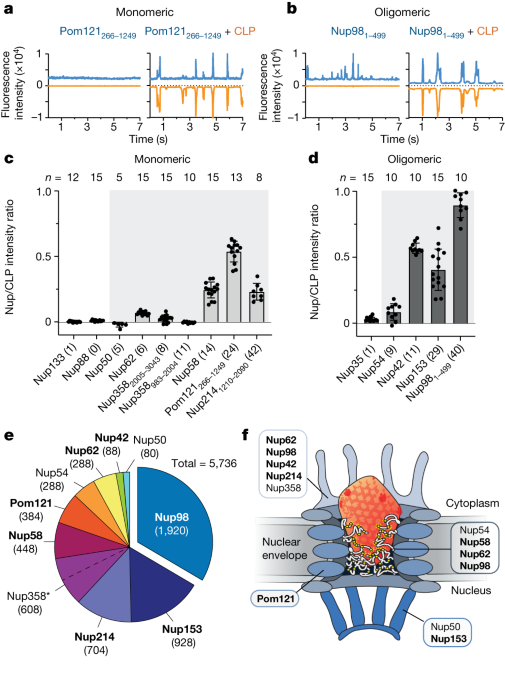
FFS was performed as described by Lau et al.17. Briefly, cell-free expressed GFP-Nup proteins (approximately 50 nM; equivalent to approximately 2,500 photon count) or AF488 peptides (100 nM) were mixed with AF568-HIV-1 CA assemblies (12 μM) in 50 mM Tris, pH 8, 150 mM NaCl. Fluorescence traces were recorded for 15 s per trace in 1 ms bins using a scanning stage operated at 1 μm s−1. Typically, measurements were repeated 10–15 times per sample.
Determining the number of FGs in relative solvent accessibility
FG motif accessibility was determined by calculating per-residue relative solvent accessibility (RSA) from AlphaFoldDB structures to determine order and disorder57. RSA-based order and disorder for all Nups except Nup358 were accessed through Mobi-DB58.
Owing to the length of Nup358, AlphaFold2 predictions were performed as three FG-containing sections (982–2004, 2005–3043 and 3058–3224), with their per-residue RSA-based disorder propensity calculated locally. Binary designations of order and disorder were assigned using a threshold optimized on Critical Assessment of Protein Intrinsic Disorder (CAID) data (0.581)57.
Phase separation
For phase-separation assays, the Nup98-saturated Ni slurry (‘Protein Expression and Purification’; Nup98 binding capacity Ni-beads 40 mg ml−1 medium) was mixed with Alexa Fluor 488-C5-maleimide (AF488, Thermo Fisher Scientific; final concentration 10 µM), followed by incubation for 5 min. We aimed for as little labelling with AF488 as possible with enough signal to noise to avoid confounding effects from the fluorophore for phase separation. The slurry was washed with 10 column volumes of wash buffer 2 (500 mM GuHCl, 20 mM imidazole pH 8) + 2 mM DTT, followed by a second wash step with wash buffer 2 without DTT to remove the DTT and residual fluorophore. The labelled protein was eluted with elution buffer for a final protein concentration of approximately 4 mg ml−1 (estimated from sodium dodecyl sulfate polyacrylamide gel electrophoresis against a bovine serum albumin standard), and phase separation of Nup98 was induced by 1:10 shock dilution into assay buffer (50 mM Tris pH 7.5, 150 mM NaCl). We observed the most ‘fluid’ condensates using shock dilution; these condensates ‘hardened’ within minutes of forming, as tested with an importin-β:IBB-mCherry control. This resulted in limited equilibration of the condensates with some fluorescent substrates (for example, as seen for CAhexamer-mCherry). Fluorescent substrates were added to the phase-separated Nup98 condensates and immediately imaged.
Confocal microscopy
Imaging was performed with a Zeiss LSM 880 inverted laser scanning confocal microscope using a ×63 oil immersion objective (numerical aperture = 1.4) (Leica). The substrate–Nup98 reaction mixes were transferred into a 12-well silicone chamber (Ibidi) on a 170 ± 5 µm cover slide. Z-stacks were taken around the centres of the phase-separated Nup98 condensates (position with highest diameter) with sequential imaging at 488 and 568. Images were processed using ImageJ2 v.2.9.0/1.53t, MATLAB R2020a and Prism v.9.4.1.
Nup98 condensate-CA experiments were performed with mCherry-labelled CA (CA-mCherry and CAhexamer-mCherry), as it has previously been reported that fluorescent dyes such as Alexa568 can non-specifically interact with Nup condensates59.
Experimental details for Fig. 3a–f and Extended Data Figs. 6 and 7: Nup98 condensates were immediately mixed with the following proteins: mCherry and IBB-mCherry (200 µM), importin-β:IBB-mCherry and importin-β + mCherry (both 100 µM), CA-mCherry and CA N57A-mCherry (45 µM), CAhexamer-mCherry and CA N57Ahexamer-mCherry (65 µM). The mixed Nup98–protein samples were imaged after around 5 min incubation on the coverslip.
Experimental details for Fig. 3h,i and Extended Data Figs. 8–10: for the CPSF6P preincubation sample, CA-mCherry, CAhexamer-mCherry and CA N74Dhexamer-mCherry (65 µM final) were incubated with CPSF6 (500 µM final) for 10 min, followed by mixing with Nup98 condensates and imaging after around 5 min incubation on the coverslip. The control sample and the CPSF6 postincubation sample were incubated with MQW for 10 min, followed by mixing with Nup98 condensates and imaging after around 5 min incubation on the coverslip. For the CPSF6 postincubation sample, CPSF6 was added to the sample on the coverslip and imaged after 5, 10, 15 and 20 min.
Experimental details for Extended Data Fig. 8c–j: CA-mCherry and CAhexamer-mCherry (final 25 µM) were incubated with importin-α:importin-β, TNPO1 and TNPO3 (all final 2.5 µM), respectively, for 10 min, followed by mixing with Nup98 condensates and imaging after around 5 min incubation on the coverslip.
Experimental details for Fig. 4 and Extended Data Fig. 9b–d: CLP-mCherry was assembled with a ratio of 1:100 CA-mCherry (final 0.4 µM) to CA (final 40 µM) by addition of NaCl (final 1 mM) and 15 min incubation at 37 °C. After assembly, the sample was spun down hard (18,000g, 7 min) to separate the assemblies from non-assembled monomer. The pellet was resuspended in assay buffer (50 mM Tris pH 7.5, 150 mM NaCl; resuspension volume the same as the original sample volume), followed by a slow spin of the resuspended pellet (4,000g, 5 min) to remove aggregates. Nup98 condensates were immediately mixed with CA-mCherry assemblies (1 µM monomer concentration) and imaged after around 5 min incubation on the coverslip.
Radial intensity profiles
Averaged radial intensity profiles were obtained using the plugin Radial Profile in ImageJ. Z-slices were chosen at the centres of the phase-separated Nup98 condensates (position with largest diameter). Profiles were background subtracted. The edge of a condensate was defined from the 488 channel as the first intensity value greater than 5. Then, 200 nm was subtracted from this value to account for point spread function of the fluorescent pixel. This point was defined as point 0 µm in the radial averaged intensity graphs (Fig. 3 and Extended Data Figs. 6–8).
Cryo-electron microscopy
Nup98 condensates were prepared as for confocal microscopy with the following modifications. To induce the formation of smaller condensates, the unlabelled Nup98 was diluted two-fold in 500 mM GuHCl, 500 mM imidazole pH 8, before a 1:10 shock dilution into assay buffer (50 mM Tris pH 7.5, 150 mM NaCl).
Unlabelled CLPs were prepared as described above. A two-step centrifugation protocol was performed as for confocal microscopy. Equal volumes of CLP suspension were mixed with freshly shock-diluted condensates, immediately before to plunge freezing to minimize sample aggregation.
Frozen-hydrated sample preparation
A mixture (4.5 µl) of freshly mixed Nup98 condensate and capsid with protein A-gold (10 nm) was applied onto a glow-discharged Quantifoil R2/2 copper grid (Quantifoil Micro Tools). The grid was blotted in the front for 2.5 s at 15 °C with 90% relative humidity and then plunged into liquid ethane using a Lecia EM GP device (Lecia Microsystem). The vitrified grids were then stored in liquid nitrogen before cryo-ET imaging.
Cryo-ET imaging and reconstruction
The grids were imaged on a Talos Arctica electron microscope (Thermo Fisher Scientific) operated at 200 kV acceleration voltage. Cryo-ET data were collected with single-axis tilt on a Falcon III direct electron detector (Thermo Fisher Scientific) in linear mode at a magnification of ×28,000 with a pixel size of 5.23 Å. Tilt series were collected using the dose-symmetric scheme60 from −60 to 60° at 3° intervals using Tomography software (Thermo Fisher Scientific) with the defocus value set to −10 µm. Total dose for each tilt series ranged from 60–70 e/Å2. Images of tilt series were binned two-fold before tomograms were reconstructed. Three-dimensional reconstructions from tilt series were generated with the IMOD package61. Fiducial tracking was used to align the stack of tilted images.
Statistics and reproducibility
Confocal microscopy images and graphs in Fig. 3 and Extended Data Figs. 6–8 show representative data from at least three independent experiments with similar results. Confocal microscopy images in Fig. 4a and Extended Data Fig. 9 are representative of three independent experiments with similar results. Cryo images and graphs in Fig. 4b–h and Extended Data Fig. 9 show representative data from two independent experiments with similar results.
Reporting summary
Further information on research design is available in the Nature Portfolio Reporting Summary linked to this article.